Lysosomal Damage Responses Underling Protein Lifetime Regulation

Project Associate Professor, Osaka University graduate School of medicine
https://en.yoshimori-lab.com/
researchmap: https://researchmap.jp/map3
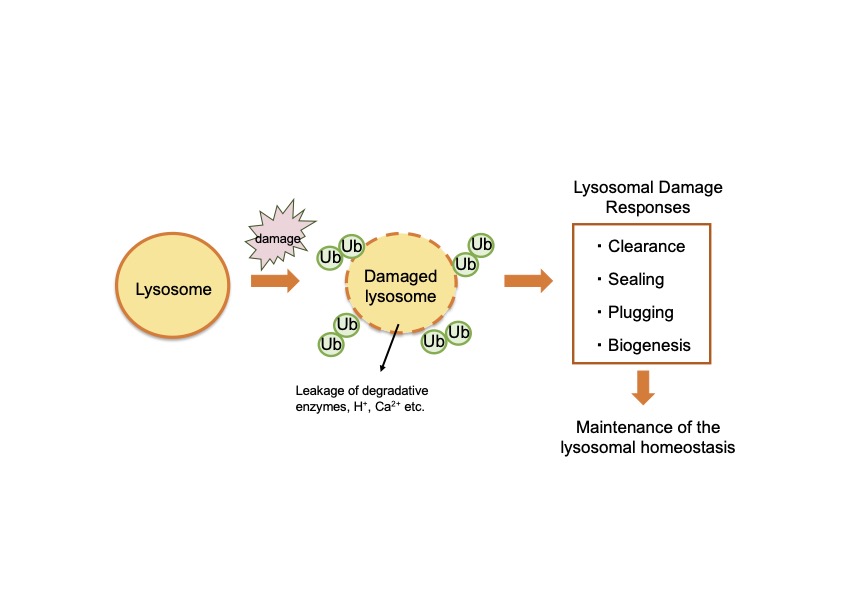
Lysosomes are organelles suitable for bulk proteolysis and are thought to play a major role when the protein composition are drastically reconfigured. Therefore, it is important to maintain lysosomal homeostasis. Recently, it has been reported that various lysosomal damage responses occur when the lysosomal membrane is damaged, but the relationship between each pathway and the details of the molecular mechanisms involved are not well understood. In this study, focusing on selective autophagy for damaged lysosomes (lysophagy), we will elucidate the molecular mechanisms of the lysosomal damage responses. In addition, lysosomes are highly ubiquitinated upon damage, but the actual significance of this process is not well understood. The involvement of multiple E2 (ubiquitin-binding enzymes) and E3 (ubiquitin ligases) has been previously reported, suggesting that proteome remodeling occurs on damaged lysosomes. In this study, we also analyze the significance of proteome remodeling on damaged lysosomes. Through these analyses, we aim to understand the mechanisms that maintain lysosomal homeostasis, which underlies protein lifetime regulation.
- Kitta S., Kaminishi T., Higashi M., Shima T., Nishino K., Nakamura N., Kosako H., Yoshimori T., & Kuma A. (2024) YIPF3 and YIPF4 regulate autophagic turnover of the Golgi apparatus. EMBO J. 10.1038/s44318-024-00131-3
- Kakuda K., Ikenaka K., Kuma A., Doi J., Aguirre C., Wang N., Ajiki T., Choong C.-J., Kimura Y., Badawy S. M. M., Shima T., Nakamura S., Baba K., Nagano S., Nagai Y., Yoshimori T., & Mochizuki H. (2024) Lysophagy protects against propagation of α-synuclein aggregation through ruptured lysosomal vesicles. Proc Natl Acad Sci U S A. 121, e2312306120
- Shima T., Ogura M., Matsuda R., Nakamura S., Jin N., Yoshimori T., & Kuma A. (2023) The TMEM192-mKeima probe specifically assays lysophagy and reveals its initial steps. J Cell Biol. 10.1083/jcb.202204048
- Saito T., Kuma A., Sugiura Y., Ichimura Y., Obata M., Kitamura H., Okuda S., Lee H.-C., Ikeda K., Kanegae Y., Saito I., Auwerx J., Motohashi H., Suematsu M., Soga T., Yokomizo T., Waguri S., Mizushima N., & Komatsu M. (2019) Autophagy regulates lipid metabolism through selective turnover of NCoR1. Nat Commun. 10, 1567
- Yoshii S. R., Kuma A., Akashi T., Hara T., Yamamoto A., Kurikawa Y., Itakura E., Tsukamoto S., Shitara H., Eishi Y., & Mizushima N. (2016) Systemic Analysis of Atg5-Null Mice Rescued from Neonatal Lethality by Transgenic ATG5 Expression in Neurons. Dev Cell. 39, 116–130
