Analysis of nutritional response and adaptation mechanism of cells based on comprehensive measurement of protein half-life

Professor, Graduate School of Science, Nagoya City University
http://www.nsc.nagoya-cu.ac.jp/profile/nakatsukasa.html
researchmap: https://researchmap.jp/read00672_0_9/?lang=japanese
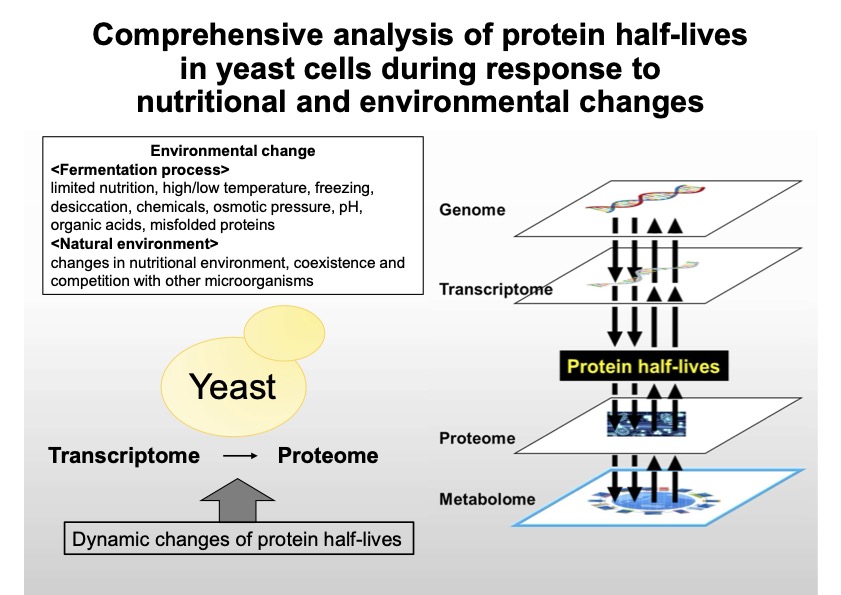
Microorganisms are exposed to rapidly changing nutritional environments during fermentation production processes and in the natural world. The response mechanism to changes in the nutritional environment has been analyzed focusing on mRNA (transcription) and protein (translation) levels, but the correlation between them is not always high. To elucidate the overall picture of the nutrient response mechanism, it is imperative to understand the post-translational regulation, especially the rapid and precise regulation by proteolysis. In this study, we will analyze protein lifetimes (half-lives) in budding yeast to understand changes in response to nutritional changes using the dynamic SILAC method. At the same time, we will follow these systematic results with a conventional analysis that delves deeply into the degradation pathways of individual proteins and the significance of degradation. This research is expected to elucidate the basic biological problem of survival strategies of microorganisms in nature and simultaneously provide fundamental knowledge toward the creation of microorganisms with environmental resistance advantageous for industrial use.
- Ikeda T., Yamazaki K., Okumura F., Kamura T., & Nakatsukasa K. (2024) Role of the San1 ubiquitin ligase in the heat stress-induced degradation of nonnative Nup1 in the nuclear pore complex. Genetics. 10.1093/genetics/iyae017
- Nishio K., Kawarasaki T., Sugiura Y., Matsumoto S., Konoshima A., Takano Y., Hayashi M., Okumura F., Kamura T., Mizushima T., & Nakatsukasa K. (2023) Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress. Sci Adv. 9, eadf1956
- Kusama K., Suzuki Y., Kurita E., Kawarasaki T., Obara K., Okumura F., Kamura T., & Nakatsukasa K. (2022) Dot6/Tod6 degradation fine-tunes the repression of ribosome biogenesis under nutrient-limited conditions. iScience. 25, 103986
- Nakatsukasa K., Wigge S., Takano Y., Kawarasaki T., Kamura T., & Brodsky J. L. (2022) A positive genetic selection for transmembrane domain mutations in HRD1 underscores the importance of Hrd1 complex integrity during ERAD. Curr Genet. 68, 227–242
- Nakatsukasa K., Nishimura T., Byrne S. D., Okamoto M., Takahashi-Nakaguchi A., Chibana H., Okumura F., & Kamura T. (2015) The Ubiquitin Ligase SCF(Ucc1) Acts as a Metabolic Switch for the Glyoxylate Cycle. Mol Cell. 59, 22–34
