Comprehensive identification and lifetime analysis of endoplasmic reticulum-associated degradative substrates in embryogenesis.

Laboratory Head, Tokyo Metropolitan Institute of Medical Science
https://www.igakuken.or.jp/pro-meta/index-e.html
researchmap: https://researchmap.jp/read0000903
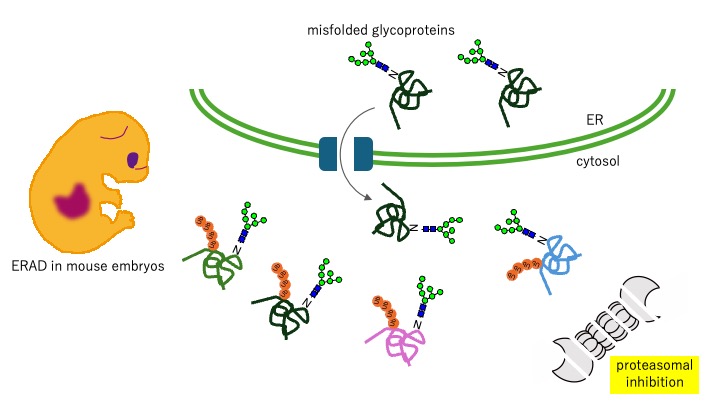
Endoplasmic reticulum-associated degradation (ERAD) is a crucial intracellular quality control system by the ubiquitin-proteasome system. Deletion of key components of ERAD results in embryonic lethality in mice, underscoring the physiological significance of ERAD in development. Although the ERAD-related molecules responsible for the execution of ERAD have been elucidated in considerable detail, most analyses of this mechanism have been performed using model proteins with pathogenic mutations. It is unclear how many endogenous proteins without mutations are degraded by the ERAD pathway. Not only misfolded proteins, but also constitutively substrates degraded by the ERAD have been reported, and this pathway is now understood as a system that actively regulates the proteome.
In this study, we comprehensively identify ubiquitinated glycoproteins from mouse embryos in which proteasome activity is attenuated by deletion of NGLY1, thereby providing a proteome-wide understanding of ERAD, which is important in development. We aim to understand intrinsic signal of substrates for ERAD by analyzing the correspondence between identified substrates and ERAD targeting molecules.
- Yoshida Y., Asahina M., Murakami A., Kawawaki J., Yoshida M., Fujinawa R., Iwai K., Tozawa R., Matsuda N., Tanaka K., & Suzuki T. (2021) Loss of peptide: N -glycanase causes proteasome dysfunction mediated by a sugar-recognizing ubiquitin ligase. Proceedings of the National Academy of Sciences. 10.1073/pnas.2102902118
- Yoshida Y., & Tanaka K. (2018) Cytosolic N-Glycans: Triggers for Ubiquitination Directing Proteasomal and Autophagic Degradation: Molecular Systems for Monitoring Cytosolic N-Glycans as Signals for Unwanted Proteins and Organelles. BioEssays. 10.1002/bies.201700215
- Yoshida Y., Yasuda S., Fujita T., Hamasaki M., Murakami A., Kawawaki J., Iwai K., Saeki Y., Yoshimori T., Matsuda N., & Tanaka K. (2017) Ubiquitination of exposed glycoproteins by SCFFBXO27 directs damaged lysosomes for autophagy. Proc Natl Acad Sci U S A. 114, 8574–8579
- Yoshida Y., Saeki Y., Murakami A., Kawawaki J., Tsuchiya H., Yoshihara H., Shindo M., & Tanaka K. (2015) A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc Natl Acad Sci U S A. 112, 4630–4635
- Kumanomidou T., Nishio K., Takagi K., Nakagawa T., Suzuki A., Yamane T., Tokunaga F., Iwai K., Murakami A., Yoshida Y., Tanaka K., & Mizushima T. (2015) The structural differences between α glycoprotein specific F-box protein Fbs1 and its homologous protein FBG3. PLoS One. 10.1371/journal.pone.0140366
