Elucidation of endogenous defective proteins using newly-developed probes.

Professor, School of Science, Tokyo Metropolitan University
https://www.biol.se.tmu.ac.jp/en-labo.asp?ID=en-celche
researchmap: https://researchmap.jp/hkawa/
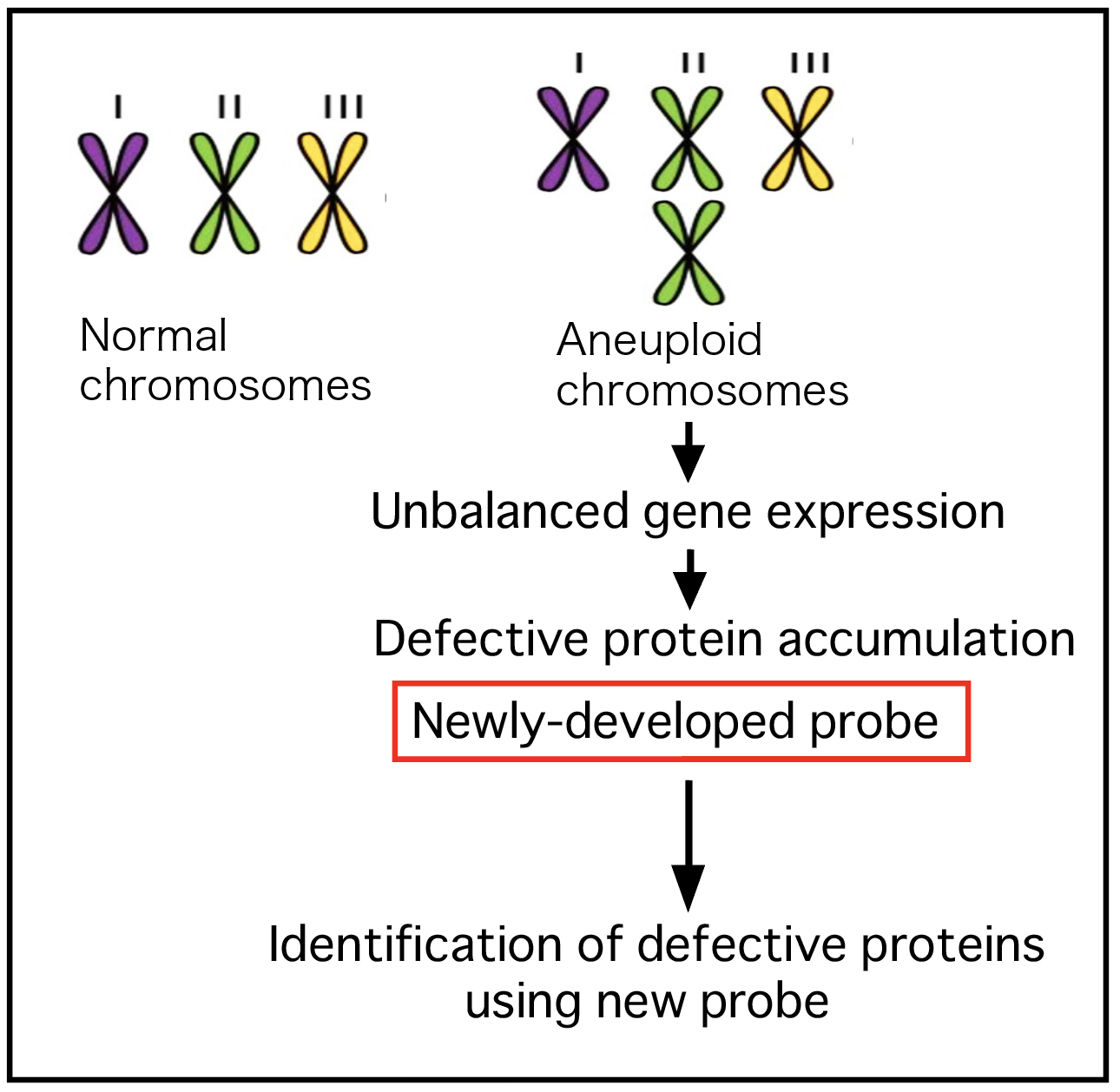
The accumulation of defective polypeptides is a major cause of various diseases. However, diversity of defective proteins and their instability are major obstacles in identifying defective polypeptides. Therefore, it is imperative to develop a probe that allows for the retrieval of defective proteins. We previously identified BAG6 as a recognition factor for exposed hydrophobicity of defective proteins (Kikukawa et al., FEBS J. 2005; Minami et al., J. Cell Biol. 2010; Suzuki & Kawahara, EMBO Rep. 2016). Utilizing our knowledge about the substrate recognition domain of BAG6 (Kikukawa et al., FEBS J. 2005; Tanaka et al., FEBS J. 2016), we constructed a novel protein architecture which is useful for detecting a variety of defective proteins in cells. We also established important negative control to test the specificity. With this probe, we are going to isolate and quantify endogenous aberrant proteins.
- Miyauchi M., Matsumura R., & Kawahara H. (2023) BAG6 supports stress fiber formation by preventing the ubiquitin-mediated degradation of RhoA. Mol Biol Cell. 10.1091/mbc.E22-08-0355
- Takahashi T., Minami S., Tsuchiya Y., Tajima K., Sakai N., Suga K., Hisanaga S., Ohbayashi N., Fukuda M., & Kawahara H. (2019) Cytoplasmic control of Rab family small GTP ases through BAG 6 . EMBO Rep. 10.15252/embr.201846794
- Suzuki R., & Kawahara H. (2016) UBQLN 4 recognizes mislocalized transmembrane domain proteins and targets these to proteasomal degradation . EMBO Rep. 17, 842–857
- Minami R., Hayakawa A., Kagawa H., Yanagi Y., Yokosawa H., & Kawahara H. (2010) BAG-6 is essential for selective elimination of defective proteasomal substrates. Journal of Cell Biology. 190, 637–650
