Mechanism of how the proteasome determines protein lifetime

Division of Protein Metabolism, The Institute of Medical Science, The University of Tokyo
https://www.cbms.k.u-tokyo.ac.jp/lab/saeki.html
researchmap: https://researchmap.jp/myportal_saeki-ys
The proteasome determines the lifespan of a wide range of proteins by timely and selective proteolysis, but the detailed molecular mechanisms of this process remain still elusive, particularly the molecular details that identify the structural diversity of ubiquitin modifications and substrate degrons. We have found that several shuttle molecules that transport ubiquitylated substrates to the proteasome, as well as the ubiquitin-selective ATPase p97, play crucial roles in proteasomal degradation, and that under stress the shuttle molecules undergo liquid-liquid phase separation with the ubiquitylated substrates to form proteasome condensates. In this planned research, we will expand our knowledge of the molecular mechanism of substrate selection after the ubiquitylation step and the spatio-temporal control of proteasomal degradation using large-scale protein lifetime measurements and targeted protein degradation methods developed in this research area. Then, we will extract common features of proteasome substrates and essential elements for proteasomal degradation by informational analysis, and further evaluate them in cells to understand the underlying principle of the regulation of protein lifetime by proteasomes.
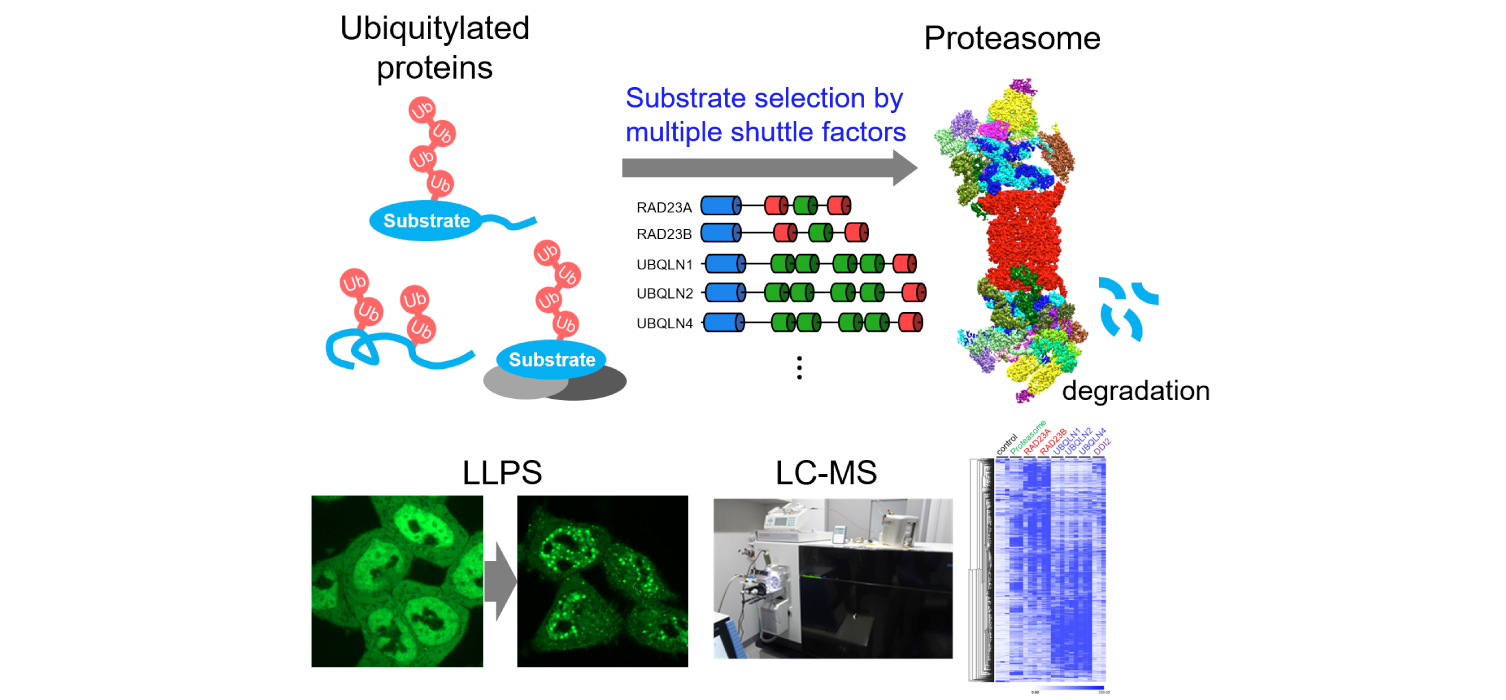
- Yasuda S, Tsuchiya H, Kaiho A, Guo Q, Ikeuchi K, Endo A, Arai N, Ohtake F, Murata S, Inada T, Baumeister W, Fernandez-Busnadiego R, *Tanaka K, *Saeki Y. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296-300 (2020)
- Sato Y, Tsuchiya H, Yamagata A, Okatsu K, Tanaka, K, *Saeki Y, *Fukai S. Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4. Nat Commun 10, 5708 (2019)
- Tsuchiya H, Burana D, Ohtake F, Arai N, Kaiho A, Komada M, Tanaka K, *Saeki Y. Ub-ProT reveals global length and composition of protein ubiquitylation in cells. Nat Commun 9, 524 (2018)
- Tsuchiya H, Ohtake F, Arai N, Kaiho A, Yasuda S, Tanaka K, *Saeki Y. In vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 axis contributes to K48-linked chain specificity of the proteasome. Mol Cell 66, 488-502 (2017)
- *Saeki Y. Ubiquitin recognition by the proteasome. J Biochem 161, 113-124 (2017)
- *Yoshida Y, Saeki Y, Murakami A, Kawawaki J, Tsuchiya H, Yoshihara H, Shindo M, *Tanaka K. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc Nat. Acad Sci USA 112, 4630-4635 (2015)
- Pack CG, Yukii H, Toh-e A, Kudo T, Tsuchiya H, Kaiho A, Sakata E, Murata S, Sako Y, Baumeister W, Tanaka K, *Saeki Y. Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat Commun 5, 3396 (2014)
- Sakata E, Bohn S, Mihalache O, Kiss P, Beck B, Nagy I, Nickell S, Tanaka K, Saeki Y, *Förster F, *Baumeister W. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc Natl Acad Sci USA 109, 1479-1484 (2012)
- Saeki Y, Toh-e A, Kudo T, Kawamura H, *Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell 137, 900-913 (2009)
- Saeki Y, Saitoh A, Toh-e A, *Yokosawa H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem Biophys Res Commun 293, 986-992 (2002)
