Protein Lifespan Dynamics in Neural Stem Cells

Graduate School of Biostudies, Kyoto University
https://takobayas.wixsite.com/homepage/index-en
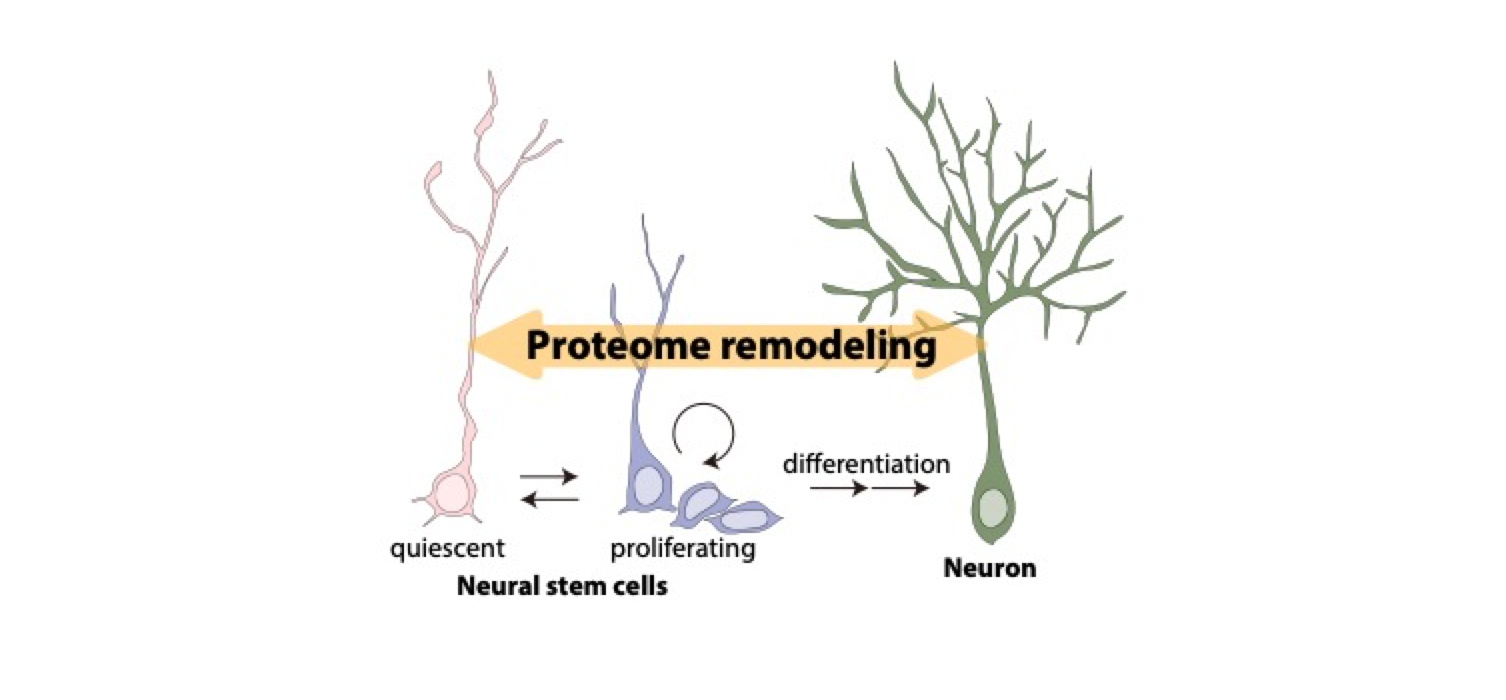
Tissue stem cells are capable of self-renewal and multipotency to differentiate into multiple cell types and maintain tissue homeostasis by supplying new functional cells. Many adult tissue stem cells enter a quiescent state, the G0/G1 phase of the cell cycle, in which proliferation and differentiation stop. Stem cells dynamically alter their properties when they are reactivated from quiescence or differentiated into functional cells. At the same time, stem cells undergo large-scale remodeling in protein composition (proteome remodeling). This is accomplished by significant changes in the proteolytic system and protein lifespan; however, the underlying mechanisms that regulate these changes remain unclear. I have previously demonstrated lysosomal regulation of neural stem cell quiescence by proteolysis of membrane receptors in lysosomes; however, the overall molecular mechanism governing this lysosomal regulation also remains unknown. In this study, I will comprehensively analyze protein lifespan during neural stem cell quiescence, activation, and differentiation to elucidate the underlying mechanisms of proteome remodeling in response to functional and state changes in neural stem cells. I will also analyze the proteome change in neural stem cell dysfunction caused by aging and brain diseases to elucidate the molecular mechanisms that control their maintenance in the brain.
- *Kobayashi T and Kageyama R. Lysosomes and signaling pathways for maintenance of quiescence in adult neural stem cells. FEBS J 288, 3082-3093 (2021)
- Zhang J, Uchiyama J, Imami K, Ishihama Y, Kageyama R and *Kobayashi T. Novel roles of small extracellular vesicles in regulating the quiescence and proliferation of neural stem cells. Front Cell Dev Biol 9, 762293 (2021)
- *Kobayashi T, Piao W, Takamura T, Kori H, (6名), Ballabio A and *Kageyama R. Enhanced lysosomal degradation maintains the quiescent state of neural stem cells. Nat Commun 10, 5446 (2019)
- *Kobayashi T, Iwamoto Y, Takashima K, Isomura A, Kosodo Y, Kawakami K, Nishioka T, Kaibuchi K and *Kageyama R. Deubiquitinating enzymes regulate Hes1 stability and neuronal differentiation. FEBS J. 282, 2475-2487 (2015)
- *Kobayashi T and Kageyama R. Expression dynamics and functions of Hes genes in development and diseases. Curr Top Dev Biol 110, 263-283 (2014)
- *Kobayashi T and Kageyama R. Hes1 oscillations contribute to heterogeneous differentiation responses in embryonic stem cells. Genes 2, 219-228 (2011)
- *Kobayashi T and Kageyama R. Hes1 oscillation: making variable choices for stem cell differentiation. Cell Cycle 9, 207-208 (2010)
- *Kobayashi T and *Kageyama R. Hes1 regulates embryonic stem cell differentiation by suppressing Notch signaling. Genes Cells 15, 689-698 (2010)
- *Kobayashi T and Kageyama R. Dynamic advances in NF-kappaB signaling analysis. Sci Signal 2, pe47 (2009)
- *Kobayashi T, Mizuno H, Imayoshi I, Furusawa C, Shirahige K and *Kageyama R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev 23, 1870-1875 (2009)
